What is mass spectrometry?
“Mass spectrometry is an analytical tool useful for measuring the mass-to-charge ratio (m/z) of one or more molecules present in a sample. These measurements can often be used to calculate the exact molecular weight of the sample components as well. Typically, mass spectrometers can be used to identify unknown compounds via molecular weight determination, to quantify known compounds, and to determine structure and chemical properties of molecules.”
(Source: Broad Institute)
Besides measuring real world substances with a mass spectrometer, theoretical mass spectra can also be calculated. A simple calculator using the mercury7 algorithm that runs in your browser can be found here. In any case, the output is a mass spectrum.
What does a mass spectrum look like?
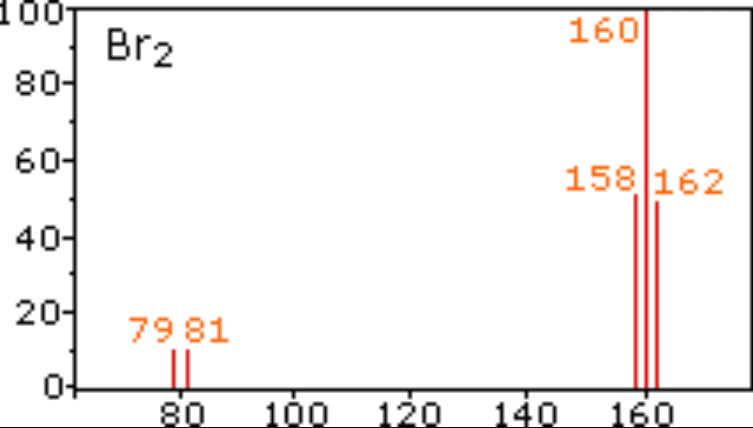
As you can see, the spectrum consists of multiple peaks that show the intensity of each m/z value detected. In a relational database this data can be stored a table rows with columns m/z and intensity, however this requires reconstruction of the spectrum from this table every time the full spectrum is required.
Alternatively, the spectrum can be stored as m/z and intensity vectors. In vector notation, the above spectrum would look like this:
intensity: [10.0, 10.0, 50.0, 100.0, 48.0]
m/z: [79.0, 81.0, 158.0, 160.0, 162.0]
Modern high precision mass spectrometers can measure m/z down to four fractional digits, so a floating point data type must be used for storage.
What is a mass spectrum database good for anyway?
As said in the introduction, mass spectrometry is one, if not the tool to identify unknown compounds, to quantify known compounds, and to determine the structure of molecules. But it is a lot of work, and you need reference spectra to compare against.
So, there are curated databases of validated spectra available, like MassBank Japan, MassBank Europe and the NIST mass spectral libraries. Laboratories might also want to store their own libraries for future use.
However, such databases often come in their own formats and with their own retrieval software. If you need to efficiently connect spectra to other data, e.g. chemical structures or genomic data, this calls for central management and a common API.
And this is where PostgreSQL and its enormous extension ecosystem shine.
ARRAY vs. pgvector
The native way to handle vectors in PostgreSQL are ARRAYs. They are first class citizens of PostgreSQL and come with lots of functionality out of the box. But now there is pgvector, an extension for vector similarity search, originally designed to handle embeddings from ML models. Let’s see how pgvector compares to ARRAY.
Test Setup
A total of 109143 spectra were loaded from here in msp format, converted into SQLite3 by msp2db, and then loaded into the following TABLE:
CREATE TABLE massbase.spectra_index (
library_spectra_meta_id int4 NOT NULL
num_peaks int2 NULL,
mz_range numrange NULL,
mz float4[] NOT NULL,
intensity float4[] NOT NULL,
mz_vector public.vector NULL,
intensity_vector public.vector NULL);
CREATE INDEX spectra_index_num_peaks_idx ON massbase.spectra_index USING btree (num_peaks);
The source TABLE generated by msp2db looks like this:
CREATE TABLE massbase.library_spectra (
id int4 NULL,
mz float4 NOT NULL,
i float4 NOT NULL,
other text NULL,
library_spectra_meta_id int4 NOT NULL);
Since this uses the one table row per m/z-intensity pair format, some conversions had to be made.
Conversion into ARRAYs:
INSERT INTO massbase.spectra_index SELECT
library_spectra_meta_id,
array_agg(mz::numeric
ORDER BY
mz asc) as mz,
array_agg(i
ORDER BY
mz ASC)
FROM
massbase.library_spectra
GROUP BY
library_spectra_meta_id
ORDER BY
library_spectra_meta_id ASC;
Building the num_peaks value:
UPDATE massbase.spectra_index SET num_peaks = array_length(mz, 1);
Building the m/z range values:
UPDATE massbase.spectra_index SET mz_range = numrange(mz[1]::numeric, mz[num_peaks]::numeric, '[]');
And finally, building the vectors from the already existing ARRAYs:
UPDATE massbase.spectra_index SET mz_vector = mz;
UPDATE massbase.spectra_index SET intensity_vector = intensity;
Testing and Results
Testing consisted of running a cosine similarity / cosine distance query 10x against various implementations (SQL, pl/pgSQL, ARRAY in C, pgvector) and different PostgreSQL configurations (out-of-the-box install, tuned buffercache 2GB, INDEX on num_peaks, parallel query, and parallel query+INDEX).
The results were measured with EXPLAIN ANALYZE with milliseconds resolution, and the median and the average were taken.
The ARRAY query:
EXPLAIN ANALYZE SELECT
library_spectra_meta_id
FROM
massbase.spectra_index
WHERE
num_peaks = 23
AND
1.0::float8 - massbase.cosine_similarity(mz, (
SELECT
mz
FROM
massbase.spectra_index
WHERE
library_spectra_meta_id = 3025
and num_peaks = 23)) < 0.1;
The pgvector query:
EXPLAIN ANALYZE SELECT
library_spectra_meta_id
FROM
massbase.spectra_index
WHERE
num_peaks = 23
AND
mz_vector <=> (
SELECT
mz_vector
from
massbase.spectra_index
WHERE
num_peaks = 23
AND
library_spectra_meta_id = 3025) < 0.1;
Out-of-the-box install, no tuning:
Run # | ARRAY pl/pgSQL | ARRAY SQL | pgvector | ARRAY C function |
1 | 257 | 270 | 190 | 186 |
2 | 249 | 266 | 191 | 183 |
3 | 248 | 258 | 192 | 180 |
4 | 259 | 263 | 199 | 162 |
5 | 250 | 257 | 198 | 163 |
6 | 256 | 259 | 205 | 176 |
7 | 241 | 274 | 206 | 174 |
8 | 248 | 266 | 200 | 174 |
9 | 256 | 271 | 197 | 174 |
10 | 260 | 263 | 195 | 183 |
Median | 253 | 264.5 | 197.5 | 175 |
Arithmetic Mean | 252.4 | 264.5 | 197.3 | 175.5 |
Tuned buffer cache to 2 GB RAM:
Run # | ARRAY pl/pgSQL | ARRAY SQL | pgvector | ARRAY C function |
1 | 102 | 112 | 53 | 105 |
2 | 113 | 114 | 62 | 102 |
3 | 104 | 119 | 56 | 94 |
4 | 103 | 115 | 54 | 103 |
5 | 102 | 113 | 55 | 96 |
6 | 109 | 120 | 54 | 93 |
7 | 110 | 117 | 52 | 94 |
8 | 107 | 114 | 57 | 95 |
9 | 109 | 117 | 59 | 101 |
10 | 102 | 123 | 56 | 95 |
Median | 105.5 | 116 | 55.5 | 95.5 |
Arithmetic Mean | 106.1 | 116 | 55.8 | 97.8 |
Tuned buffer cache to 2 GB RAM and btree index on num_peaks column:
Run # | ARRAY pl/pgSQL | ARRAY SQL | pgvector | ARRAY C function |
1 | 53 | 55 | 4 | 5 |
2 | 61 | 64 | 4 | 5 |
3 | 46 | 56 | 4 | 5 |
4 | 49 | 55 | 6 | 4 |
5 | 46 | 57 | 4 | 4 |
6 | 49 | 58 | 4 | 4 |
7 | 49 | 64 | 4 | 6 |
8 | 50 | 60 | 3 | 7 |
9 | 55 | 60 | 5 | 4 |
10 | 47 | 63 | 4 | 4 |
Median | 49 | 59 | 4 | 4.5 |
Arithmetic Mean | 50.5 | 59 | 4.2 | 4.8 |
Tuned buffer cache to 2 GB RAM and parallel query:
Run # | ARRAY pl/pgSQL | ARRAY SQL | pgvector* | ARRAY C function* |
1 | 108 | 108 | 56 | 89 |
2 | 107 | 104 | 53 | 93 |
3 | 111 | 107 | 63 | 97 |
4 | 106 | 109 | 56 | 96 |
5 | 103 | 109 | 51 | 89 |
6 | 102 | 103 | 57 | 95 |
7 | 106 | 106 | 57 | 91 |
8 | 107 | 123 | 57 | 94 |
9 | 110 | 112 | 59 | 91 |
10 | 107 | 105 | 60 | 90 |
Median | 107 | 107.5 | 57 | 92 |
Arithmetic Mean | 106.7 | 107.5 | 56.9 | 92.5 |
* never parallelized |
Tuned buffer cache to 2 GB RAM and parallel query and btree index on num_peaks column (Spoiler: It never parallelized):
Run # | ARRAY pl/pgSQL | ARRAY SQL | pgvector | ARRAY C function |
1 | 52 | 53 | 4 | 4 |
2 | 52 | 57 | 4 | 5 |
3 | 47 | 60 | 3 | 4 |
4 | 54 | 55 | 4 | 5 |
5 | 47 | 70 | 5 | 5 |
6 | 47 | 58 | 4 | 5 |
7 | 45 | 59 | 4 | 5 |
8 | 56 | 64 | 5 | 5 |
9 | 55 | 59 | 5 | 4 |
10 | 48 | 57 | 4 | 4 |
Median | 50 | 58,5 | 4 | 5 |
Arithmetic Mean | 50.3 | 58,5 | 4.2 | 4.6 |
Test system:
CPU | Intel Core-i5-10210U (4 cores / 8 threads) |
RAM | 8 GB |
SSD | WDC PC SN730 SDBPNTY-512G-1036 (NVM Express, PCIe 3.0 x4) |
Findings
- With ARRAY, C functions outperform pl/pgSQL and SQL functions in all cases, ~1.44x at worst (253 ms : 175 ms), and ~10x at best (50ms : 5ms).
- On an unoptimized instance, full table scans dominate query runtime
- A tuned PostgreSQL server and indexing significantly accelerate queries in all cases
- Parallel queries accelerate pl/pgSQL and SQL functions.
- C function queries were never parallelized
- With a suitable INDEX present, no query was parallelized
Conclusion
The C function using ARRAY is almost as fast as pgvector, but, because of the PostgreSQL ARRAY API, it is difficult to vectorize on CPU (SIMD: MMX, SSE, AVX, NEON).
The pgvector vector type is much better suited for SIMD processing, and it is quite easy to use it in your own C functions, only #include “vector.h“ is needed. No linking against the pgvector library itself is required. So, pgvector is also a good foundation to build your own vector similarity metrics, like spectral contrast angle or hamming distance.
Repository
All code used can be found here.